Blocker: Random Effects Meta-Analysis of Clinical Trials¶
An example from OpenBUGS [38] and Carlin [13] concerning a meta-analysis of 22 clinical trials to prevent mortality after myocardial infarction.
Model¶
Events are modelled as

where 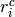 is the number of control group events, out of
is the number of control group events, out of 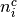 , in study
, in study 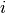 ; and
; and 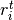 is the number of treatment group events.
is the number of treatment group events.
Analysis Program¶
using Mamba
## Data
blocker = (Symbol => Any)[
:rt =>
[3, 7, 5, 102, 28, 4, 98, 60, 25, 138, 64, 45, 9, 57, 25, 33, 28, 8, 6, 32,
27, 22],
:nt =>
[38, 114, 69, 1533, 355, 59, 945, 632, 278, 1916, 873, 263, 291, 858, 154,
207, 251, 151, 174, 209, 391, 680],
:rc =>
[3, 14, 11, 127, 27, 6, 152, 48, 37, 188, 52, 47, 16, 45, 31, 38, 12, 6, 3,
40, 43, 39],
:nc =>
[39, 116, 93, 1520, 365, 52, 939, 471, 282, 1921, 583, 266, 293, 883, 147,
213, 122, 154, 134, 218, 364, 674]
]
blocker[:N] = length(blocker[:rt])
## Model Specification
model = Model(
rc = Stochastic(1,
@modelexpr(mu, nc, N,
begin
pc = invlogit(mu)
Distribution[Binomial(nc[i], pc[i]) for i in 1:N]
end
),
false
),
rt = Stochastic(1,
@modelexpr(mu, delta, nt, N,
begin
pt = invlogit(mu + delta)
Distribution[Binomial(nt[i], pt[i]) for i in 1:N]
end
),
false
),
mu = Stochastic(1,
:(Normal(0, 1000)),
false
),
delta = Stochastic(1,
@modelexpr(d, s2,
Normal(d, sqrt(s2))
),
false
),
delta_new = Stochastic(
@modelexpr(d, s2,
Normal(d, sqrt(s2))
)
),
d = Stochastic(
:(Normal(0, 1000))
),
s2 = Stochastic(
:(InverseGamma(0.001, 0.001))
)
)
## Initial Values
inits = [
[:rc => blocker[:rc], :rt => blocker[:rt], :d => 0, :delta_new => 0,
:s2 => 1, :mu => zeros(blocker[:N]), :delta => zeros(blocker[:N])],
[:rc => blocker[:rc], :rt => blocker[:rt], :d => 2, :delta_new => 2,
:s2 => 10, :mu => fill(2, blocker[:N]), :delta => fill(2, blocker[:N])]
]
## Sampling Scheme
scheme = [AMWG([:mu], fill(0.1, blocker[:N])),
AMWG([:delta, :delta_new], fill(0.1, blocker[:N] + 1)),
Slice([:d, :s2], [1.0, 1.0])]
setsamplers!(model, scheme)
## MCMC Simulations
sim = mcmc(model, blocker, inits, 10000, burnin=2500, thin=2, chains=2)
describe(sim)
Results¶
Iterations = 2502:10000
Thinning interval = 2
Chains = 1,2
Samples per chain = 3750
Empirical Posterior Estimates:
Mean SD Naive SE MCSE ESS
delta_new -0.25005767 0.15032528 0.0017358068 0.0042970294 1223.84778
s2 0.01822186 0.02112127 0.0002438874 0.0012497271 285.63372
d -0.25563567 0.06184194 0.0007140893 0.0030457284 412.27207
Quantiles:
2.5% 25.0% 50.0% 75.0% 97.5%
delta_new -0.53854055 -0.32799584 -0.25578493 -0.17758841 0.07986060
s2 0.00068555 0.00416488 0.01076157 0.02444208 0.07735715
d -0.37341230 -0.29591698 -0.25818488 -0.21834138 -0.12842580